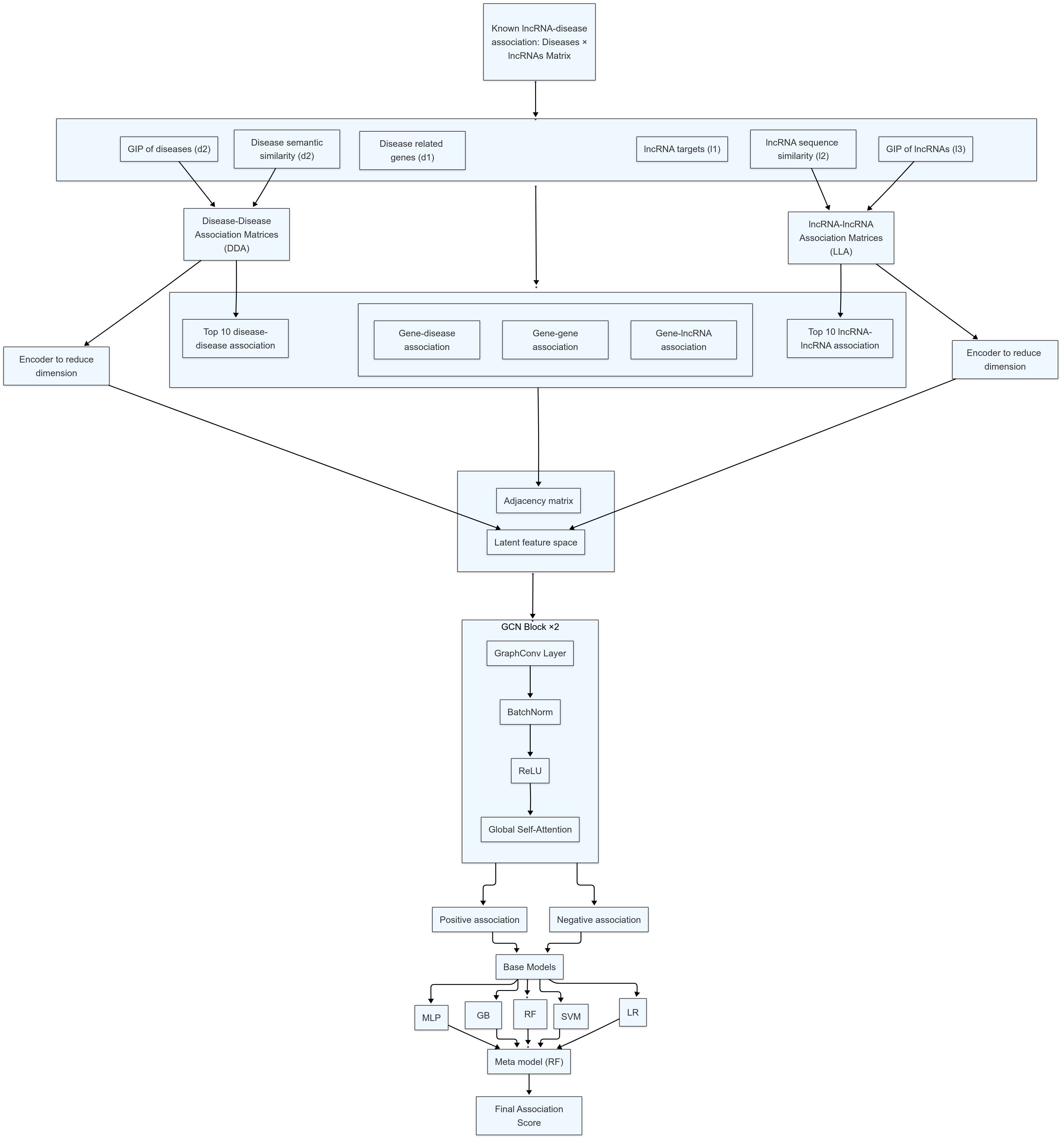
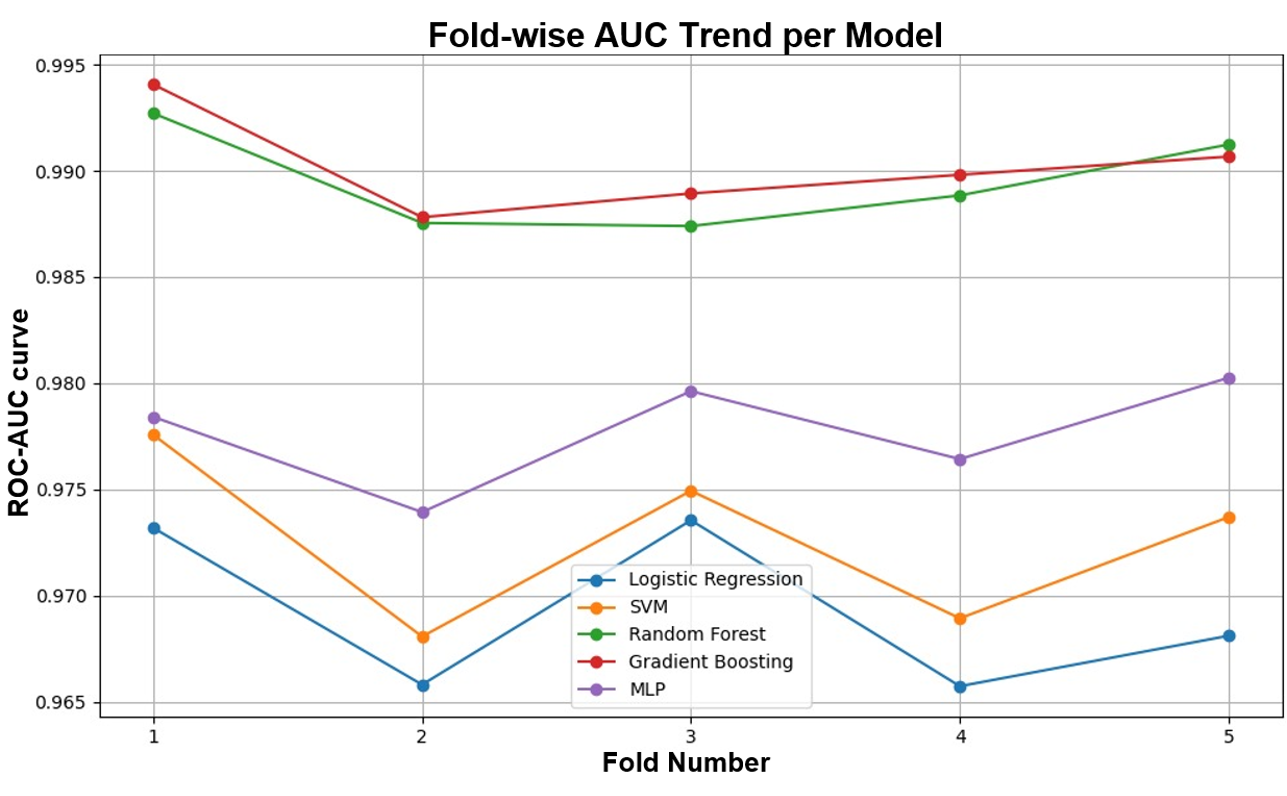
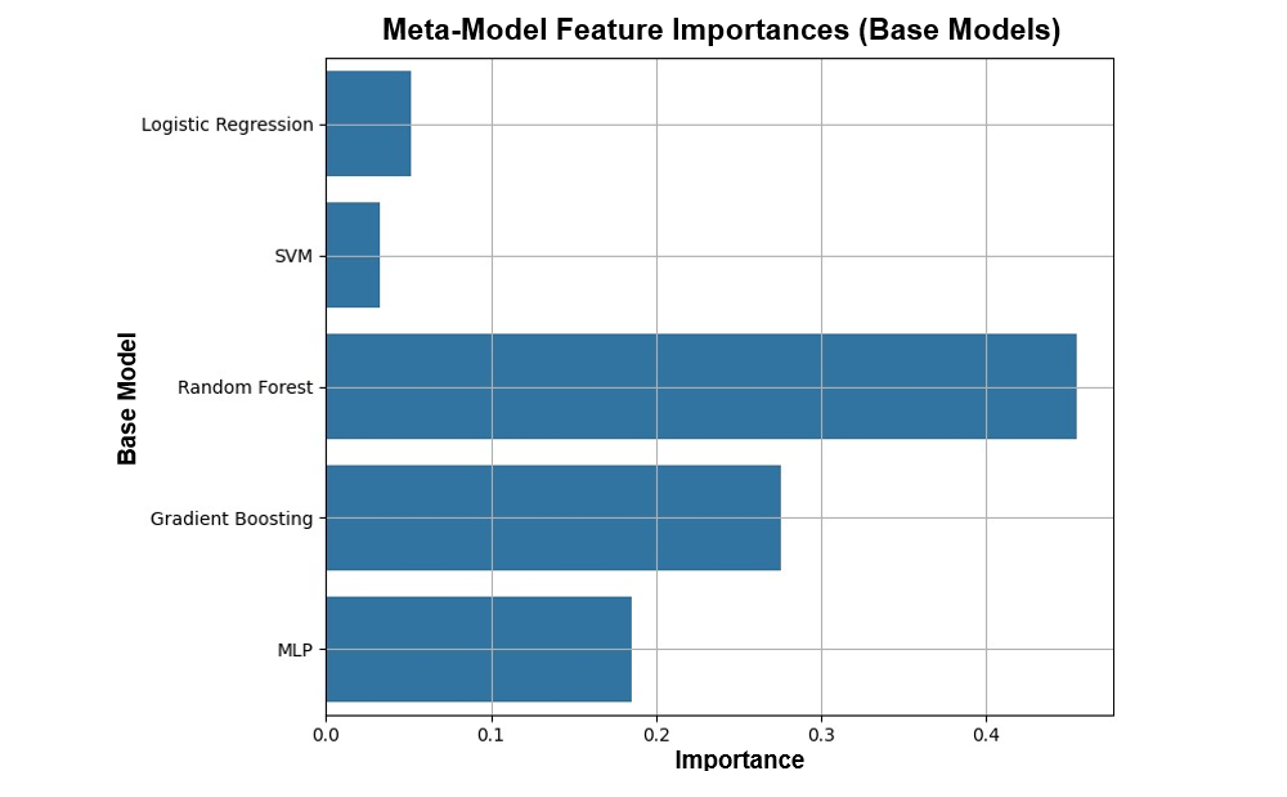
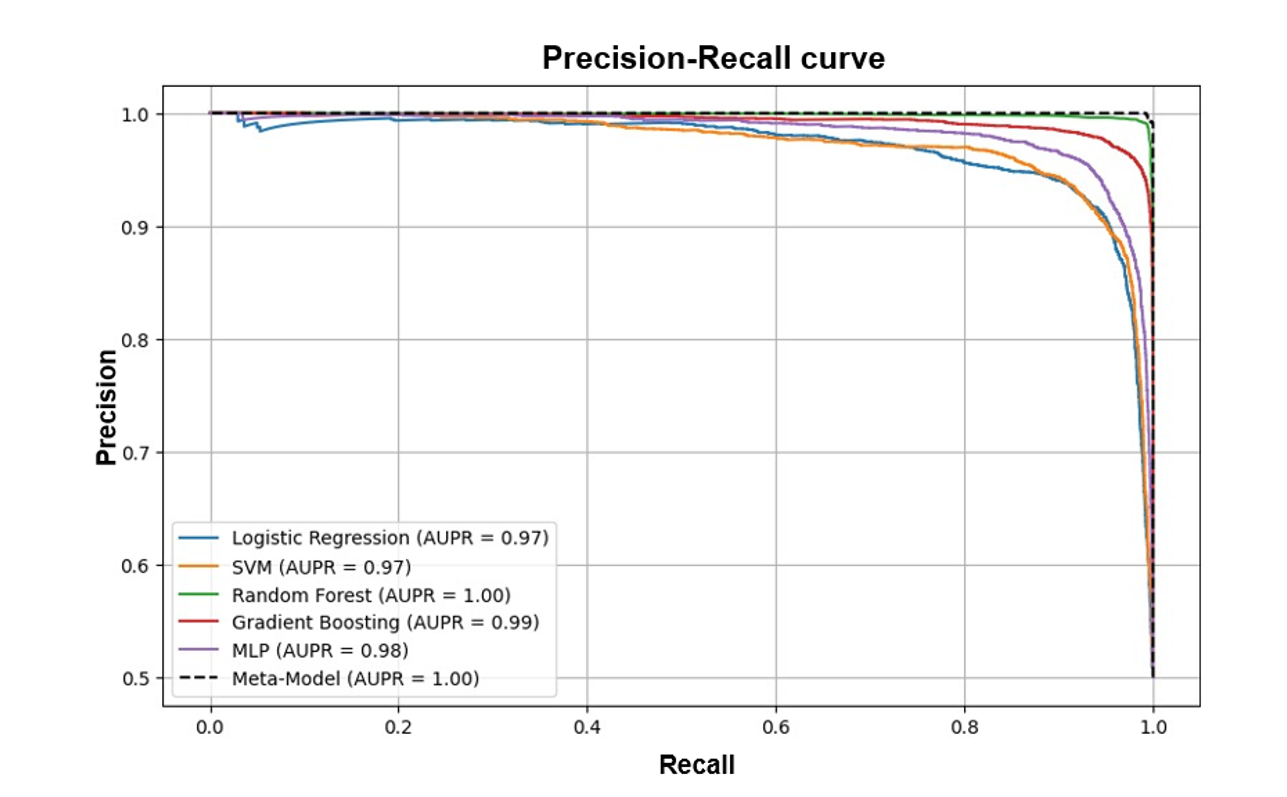
|
-
A recently discovered lncRNA, LINC0026136, gathered the attention of the research society in
2021 as found to be abnormally expressed in human tumors.
-
It primarily functions as a suppressor in cancers as well as found to be involved in
processes like motility, chemoresistance, cell proliferation, apoptosis, and tumorigenesis.
-
By conducting a manual search and a recent literature review, its role as a therapeutic
biomarker is evident. As a result, on Pubmed, we found the top 5 hits as its relation with
thyroid cancer (PMID: 34982424), colon cancer (PMID: 31850713), breast cancer (PMID:
33274565), pancreatic cancer (PMID: 32020223), and cholangiocarcinoma (PMID: 34022894).
-
Then, to evaluate the information related to LINC00261, we removed all its information from
the training data and retrained the GARNet model. After that, PrediLnc was used to extract
the top 10 associated diseases with LINC00261. The following disorders were the output:-
‘colonic neoplasms’, ‘melanoma’, ‘lung neoplasms’, ‘cholangiocarcinoma’, ‘osteoarthritis’,
‘adenocarcinoma of lung’, ‘carcinoma, renal cell’, ‘thyroid cancer, papillary’,
'nasopharyngeal carcinoma', and 'pancreatic neoplasms'. Out of 5 published top results, we
found 4 of them to be part of the top 10 diseases predicted for LINC00261.
|
-
Type 2 diabetes mellitus is a complex metabolic condition where the patient suffers from
impaired glucose regulation37.
-
Over the last five years, several lncRNAs have found a role in regulating insulin signaling,
beta cell function, inflammation resistance, and epigenetic regulation.
-
Similar activity was performed by doing a manual search on Pubmed, which shows the top 5
hits as XIST (PMID: 36628211), NEAT1 (PMID: 36027040), CASC2 (PMID: 33155514), SNHG17 (PMID:
31509021), and MALAT1 (PMID: MALAT1).
-
When we searched about type 2 diabetes mellitus using PrediLnc, then the following lncRNAs
were predicted:- ‘MALAT1’, ‘LINC00963’, ‘CASC2’, ‘WEE2-AS1’, ‘SNHG17’, ‘SNHG16’, ‘NEAT1’,
‘XIST’, ‘MIAT’, and ‘KCNQ5-AS1’. Here, we found all the top 5 hits from Pubmed were part of
the top 10 predictions by PrediLnc.
|